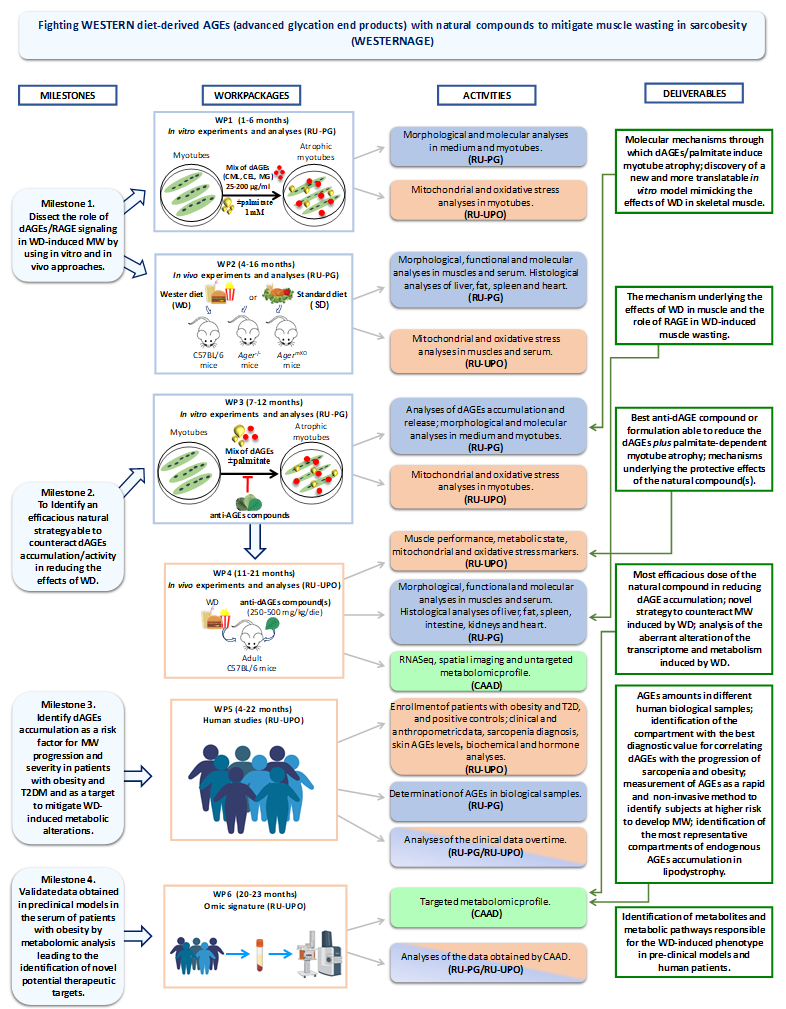
The project is subdivided in the following 6 WPs and relative Tasks to achieve 4 principal Milestones:

Methodology
WP1 In vitro experiments and analyses
C2C12 myotubes will be exposed to increasing doses of dAGE mix (25-200 μg/ml) to assess various factors at different time points, including myotube diameters, protein expressions (RAGE, MyHC), activation of the proteolytic system (atrogenes, mitophagy), mitochondrial morphology (via mitoTracker and IF), dynamics (fission and fusion markers), functionality (high-respirometry), content (mtDNA quantification), oxidative stress (ROS levels), and mt membrane polarization (JC1 staining). Pathways for protein synthesis/degradation will also be evaluated, and the role of RAGE-activated pathways will be assessed using the RAGE inhibitor FPS-MZ1.
To mimic conditions in obesity and T2D, C2C12 myotubes will be treated with a combination of dAGE mix and palmitate (1mM), a fatty acid found in Western diets. Morphological and molecular parameters will be evaluated, including the impact of palmitate alone on protein degradation, insulin resistance, and mitochondrial dysfunction.
WP2 In vivo experiments and analyses
In order to investigate the involvement of the dAGEs/RAGE axis in WD-induced effects we will feed adult C57BL/6 wild-type,
C57BL/6-Ager–/– (RAGE KO mice) and C57BL/6-AgermKO (a novel generated tamoxifen-inducible model in which the Cre
recombinase mediates RAGE gene lacking only in adult myofibers) mice (n=10, each group) with WD or SD for 18 weeks. The
availability of an inducible animal model lacking RAGE only in muscle tissue (AgermKO) mice, besides mice lacking RAGE in the
whole overall body (Ager–/–), will put at our disposal the necessary pattern of preclinical models to discover the direct role of RAGE
in WD-induced MW.
During the experiment, mice will be weighted and tested for muscle functionality (grip and Kondziela’s inverted screen test) every
three days. Food consumption will be monitored to ensure comparable food intake among the experimental groups. At the end of
the experiment, we will collect and weighted liver, heart, spleen, white and brown adipose tissues, and hind-limb skeletal muscles.
We will evaluate dAGEs expression and accumulation in muscles (WB), and serum (ELISA); muscle histology (cross-sectional area)
including the presence of adipose (red oil) and fibrotic tissues (Mallory’s trichrome stain); RAGE and fast and slow MyHC expressions
(WB); markers of proteolytic systems (WB); serum (multiplex ELISA) and muscle (real-time PCR) markers of inflammation; muscle
and serum oxidative stress (e.g., ROS detection and TBARS assay kits); mt morphology and function; histology
(hematoxylin/eosin staining) of liver, heart, spleen, adipose tissues.
WP3 In vitro experiments and analyses
The three selected anti-AGE compounds (V. macrocarpon, C. sinensis, and chlorophyll) will be added singly or mixed in group of two
(1:1) or three (1:1:1) to C2C12 myotubes treated with dAGEs and palmitate (see Task 1.2) and we will evaluate the expression of
dAGEs in myotubes (WB), dAGEs in the medium (ELISA), and the parameters like the impact of palmitate alone on protein degradation, insulin resistance, and mitochondrial dysfunction.
WP4 In vivo experiments and analyses
Mice fed with a Western diet (WD, n=10 per group) will be treated with either a vehicle or an anti-dAGEs compound (identified in WP3) at two doses (250 or 500 mg/kg/day). Control groups will consist of mice fed with a standard diet (SD). Mice will be monitored for weight, muscle function, and basal energy expenditure using metabolic cages for 72 hours before sacrifice. Afterward, organs (liver, spleen, fat, small intestine, heart, kidneys, and muscles) will be collected and analyzed. Histological examinations (hematoxylin/eosin staining) will be conducted on organs to evaluate any toxic effects of the anti-dAGEs compounds.
The gastrocnemius muscles of WD vs. SD-fed mice will undergo RNA sequencing (n=6 per group). Deregulated genes and pathways identified by Ingenuity Pathway Analysis (IPA) will be further analyzed at the protein level using Imaging Mass Cytometry (Hyperion-Helion system) to detect up to 37 proteins in muscle slices. The spatial distribution of affected proteins will be examined, and protein modulation in WD-fed mice treated with anti-dAGEs compounds will be studied using immunofluorescence (IF). Additionally, untargeted biofluid metabolomics analysis of serum samples will be performed to characterize phenotypic changes, using gas chromatography-mass spectrometry (GCxGC-MS).
WP5 Human studies
160 patients with obesity and T2D will be enrolled, with half diagnosed within the last year and half diagnosed for more than 12 months. Additionally, 35 positive controls with T2D and lipodystrophy syndrome will be included. Lipodystrophy patients typically experience insulin resistance, inflammation, sarcopenia, and mitochondrial dysfunction. These patients are part of the ECLip registry. The study protocol will be submitted to the local Ethics Committee and registered on ClinicalTrial.gov.
Patients will undergo clinical evaluations to assess body composition, including BMI, circumferences of waist, hip, arm, and calf, bioelectrical impedance analysis (BIA), dual-energy x-ray absorptiometry (DEXA), and abdominal CT or MRI scans (if applicable). Measurements will be categorized based on ESPEN/EASO consensus on sarcobesity. Patients will be classified based on complications related to T2D and its stages.
AGEs levels will be measured in biological samples (plasma, urine, skin) to identify correlations with sarcopenia progression in obesity and T2D patients. The levels of free and hemoglobin-bound AGEs, along with AGEs in urine and skin fluorescence, will be assessed. Inflammation and oxidative stress will be examined through cytokine profiles, C-reactive protein, and oxidative stress markers (ROS, TBARS). Metabolic parameters related to T2D will also be evaluated. The correlation of AGE levels, anthropometric data, muscle function, physical performance, and other clinical parameters will be analyzed over time.
WP6 Omic signature
The omic signature identified in the animal models will be validated in a selected group of patients and positive controls, i.e.,
patients (high quintile) with the highest levels of AGEs in the most representative compartment and severe sarcopenia, and patients (low quintile) with the lowest AGEs levels and stable sarcopenia by biofluid metabolomics analysis